Lei Sun (孙磊),PI
I started RNA strcuturome lab at Shandong University (Qingdao) in December of 2022. Our lab broadly interested in RNA biology, especially RNA structure biology, RNA vaccine
and adaptor design, and anti-RNA virus drug development (such as SARS-CoV-2). Our lab will develop and use a combination of computational and experimental methods to interpret the structural and functional relationships
of biological macromolecules, build the interaction network, uncover the regulation mechanism and find the potential therapy for diseases based on high-throughput sequencing, bioinformatics analysis, artificial intelligence (AI) prediction,
and experimental validation.
From 2019 to 2022, I was a Postdoc researcher at Tsinghua University, cooperating with Prof. Qiangfeng Cliff Zhang. Before my Postdoc period, I received my Ph.D. degree in the School of Life Science, Tsinghua University in 2019. Before I came to Tsinghua University, I received a bachelor's degree in Taishan College, Shandong University in 2014.
Please feel free to directly contact me via e-mail for formal or informal inquiries: sunlei0227 at sdu.edu.cn.
What's New
[Jun. 2024] Yilin Song joined our lab, welcome,Yilin!
[Jun. 2024] Yuhua Jiao joined our lab, welcome,Yuhua!
[Jun. 2024] Liangyu Li joined our lab, welcome,Liangyu!
[May 2024] Xinen Zheng joined our lab, welcome,Xin en!
[May 2024] Sen Zhang joined our lab, welcome,Sen!
[Mar. 2024] Yuning Liu joined our lab, welcome,Yuning!
[Feb. 2024] Dr. Xiaoyu Zhou joined our lab, welcome,Xiaoyu!
[Jan. 2024] Our Review paper "RNA Structure: Implications in Viral Infections and Neurodegenerative Diseases" was accepted at Advanced Biotechnology. This is the first paper produced by Sunlab only. Congratulations to Suiru, Yongkang and Shaozhen!
[Nov. 2023] Shaozhen Yin joined our lab as an intern, welcome,Shaozhen!
[Oct. 2023] Yule Gu joined our lab for graduation design, welcome,Yule!
[Oct. 2023] Long Qian joined our lab, welcome,Long!
[Sep. 2023] Dr. Jindong Sun joined our lab, welcome,Jindong!
[Sep. 2023] Yuancheng Wang joined our lab, welcome,Yuancheng!
[Sep. 2023] Yang Cheng joined our lab, welcome,Yang!
[Aug. 2023] Jingwen Wang joined our lab, welcome,Jingwen!
[Jul. 2023] Junhao Wang joined our lab, welcome,Junhao!
[Jul. 2023] Ziyou Luo joined our lab, welcome,Ziyou!
[Jun. 2023] Dr. Tong Zhou joined our lab, welcome,Tong!
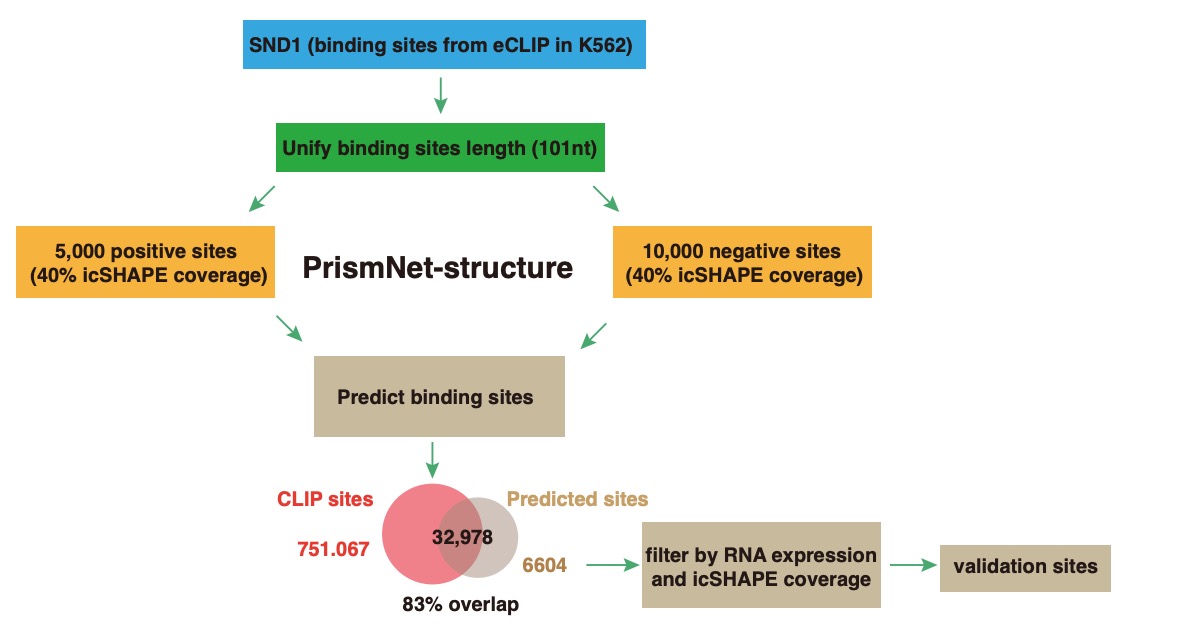
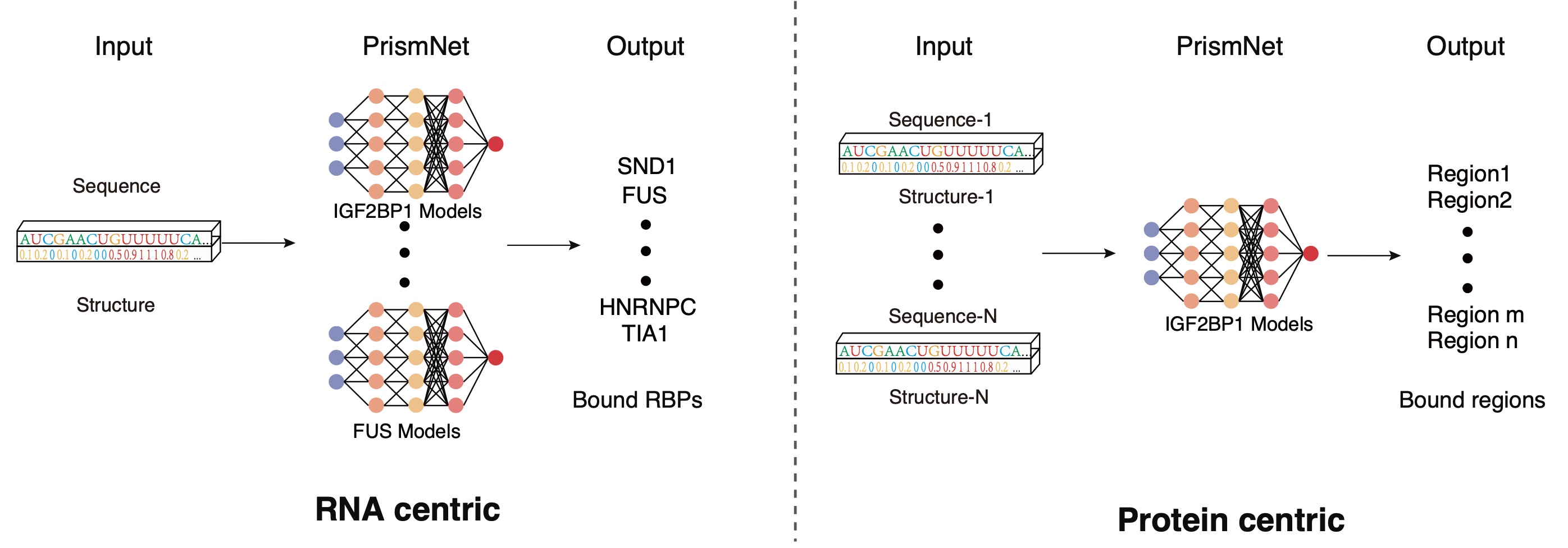
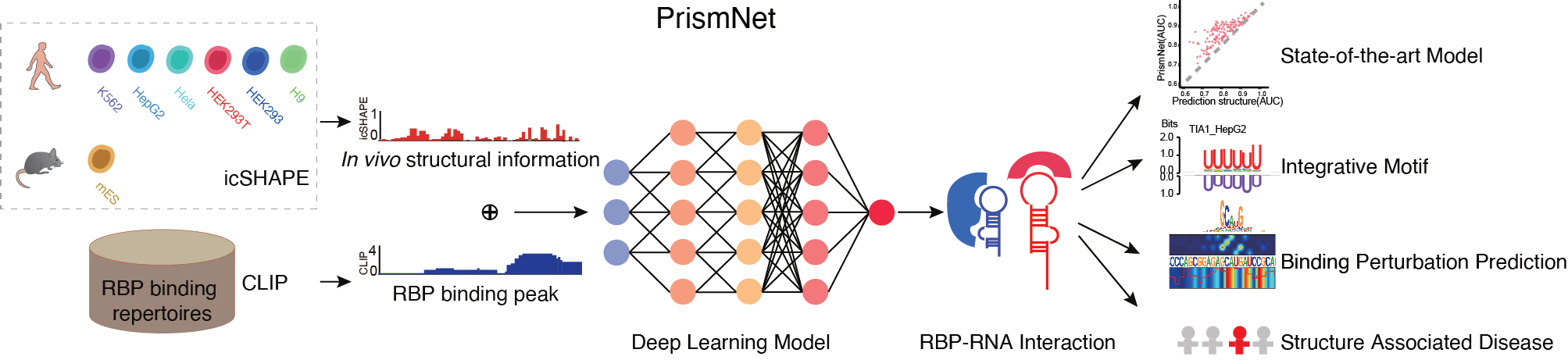
[May 2023] Our PrismNet web server paper "PrismNet: predicting protein–RNA interaction using in vivo RNA structural information" was accepted at Nucleic Acids Research. This is my first paper in Shandong University.
[May 2023] Chen Yan joined our lab, welcome,Chen!
[May 2023] Jiale He joined our lab, welcome,Jiale!
[Apr. 2023] Dr. Sun gave an invited virtual talk to the Daxue Club about RNA structurome and AI improving drug development.
[Apr. 2023] Ruobin Zhao joined our lab, welcome,Ruobin!
[Apr. 2023] Chengqian Wang joined our lab, welcome,Chengqian!
[Mar. 2023] Suiru Lu joined our lab, welcome,Suiru!
[Mar. 2023] Shanghao Dai joined our lab, welcome,Shanghao!
[Feb. 2023] Yongkang Tang joined our lab, welcome,Yongkang!
[Feb. 2023] Lingyun Xu joined our lab, welcome,Lingyun!
[Dec. 2022] Dr.Sun joined the School of Life Science, Shandong University and started the RNA structurome lab.
[Nov. 2022] Our PrismNet method paper win the 2021 Sanofi-Cell Research Outstanding Research Article Award.
[Oct. 2022] Our Review Article paper "Advances and opportunities in RNA structure experimental determination and computational modeling" was online at Nature Methods. congratulations to all the team members.
[Feb. 2022] Our PrismNet method paper was awarded as the Top Ten Advances of 2021 in Bioinformatics in China by Genomics, Proteomics and Bioinformatics (GPB)
[Jan. 2021] Our PrismNet method paper was accepted by Cell Research, congratulations to all the team members.
[Nov. 2020] Our anti-SARS-CoV-2 drugs based on RNA structures paper was accepted by the top journal Cell, congratulations to all the team members.
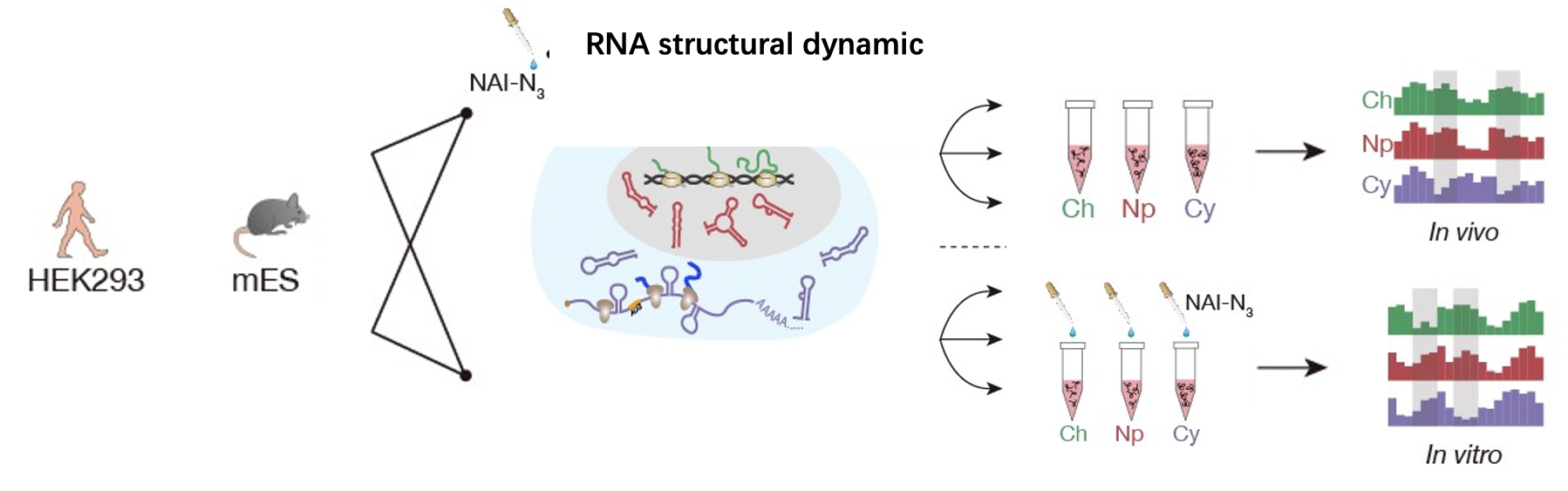
[Feb. 2019] Our RNA structural dynamics paper in different cell compartments was accepted by Nature Structural & Molecular Biology, congratulations to all the team members.
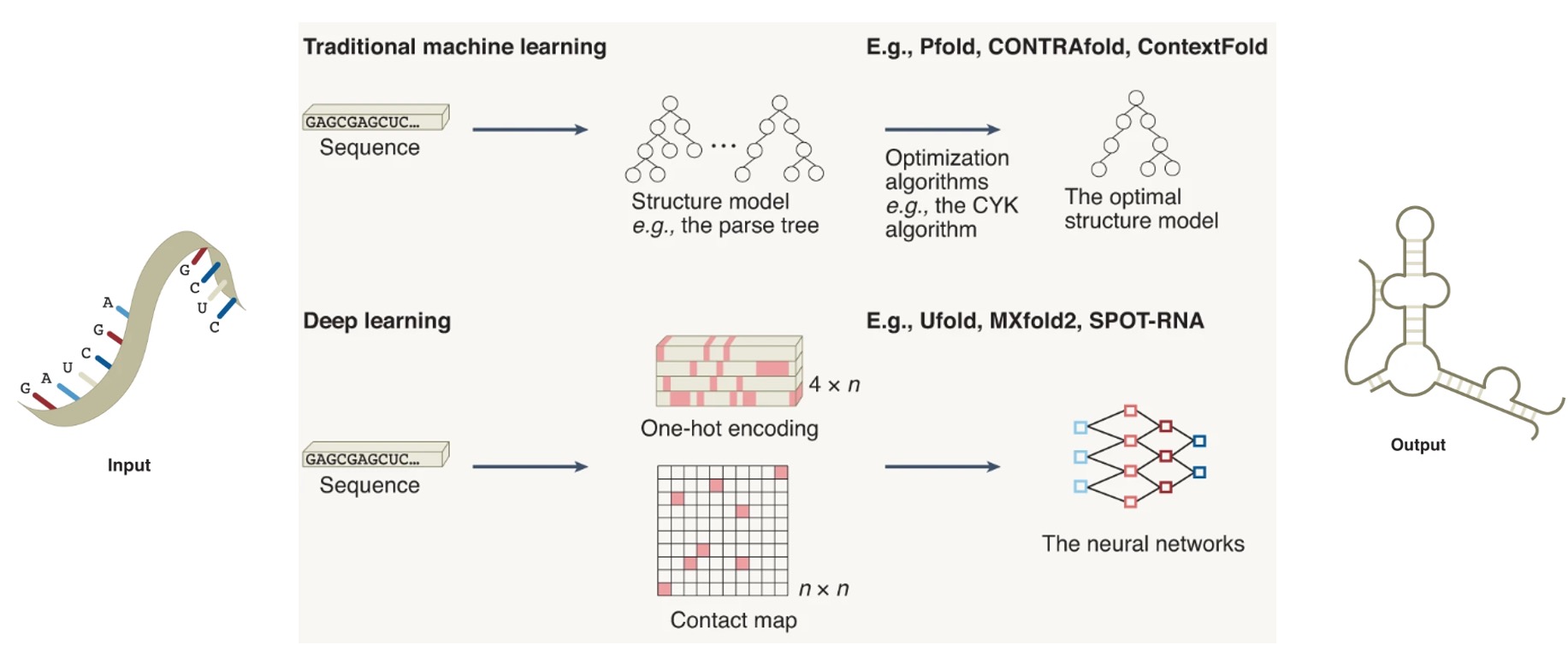
Research Interest
1. Technology development
Our lab is both interested in experimental and computational methods development. We have taken part in the development of many
technologies including subcellular RNA structure determination (Sun et.al., NSMB, 2019 )and RBP-RNA interaction prediction based
on artificial intelligence (Sun et.al., Cell Research, 2021).
We are also interested in single-cell RNA structure determination, and co-transcriptional transient RNA structure determination in the future.
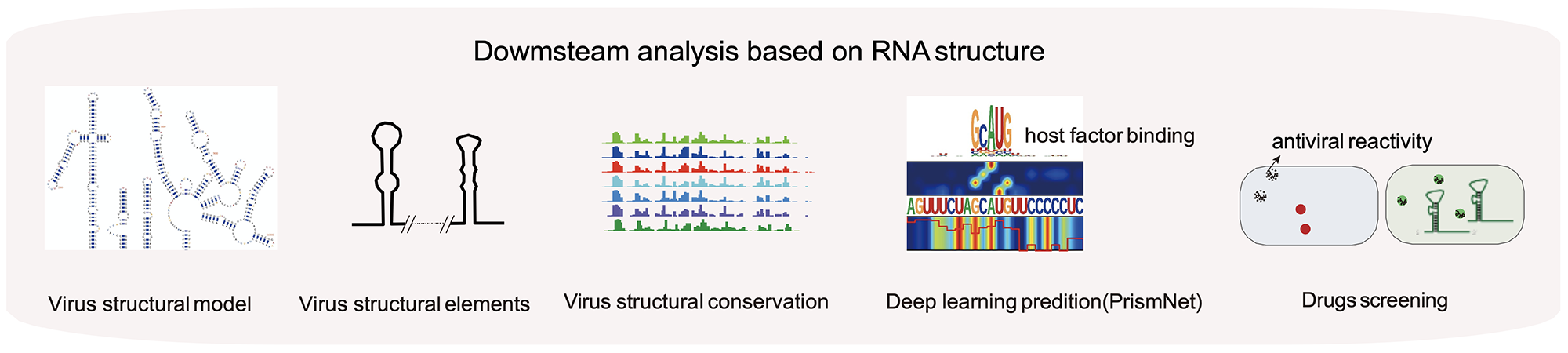
2. Biological question
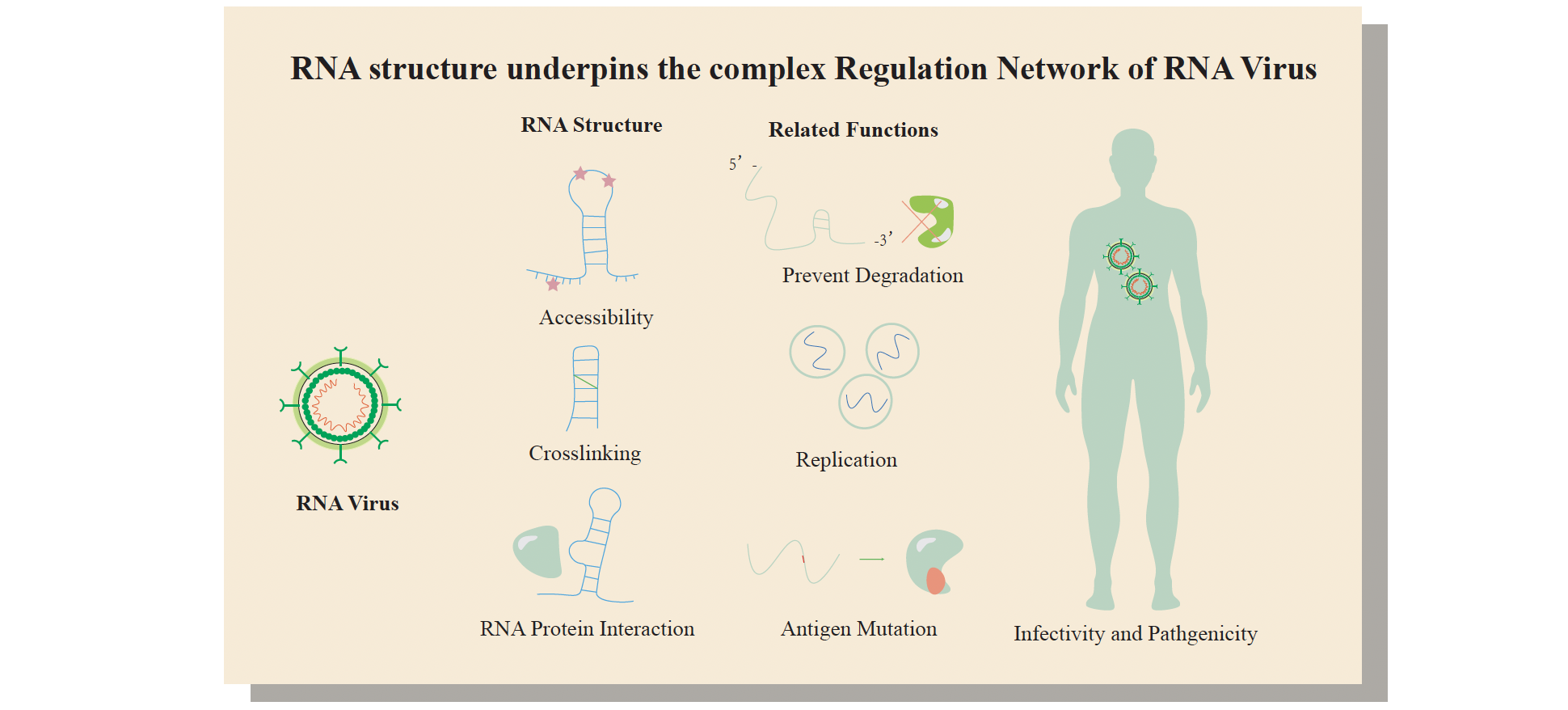
RNA structure and RBP-RNA interactions play key roles in post-transcriptional regulation. The regulatory network of viruses can be studied from the perspectives
of RNA structure and RNA-protein interaction by icSHAPE, PrismNet, CLIP, and other techniques. Antiviral drugs can be developed by targeting important structural
elements of viruses or interacting host proteins (Sun et.al., Cell, 2021).
It is important to note that the integration of high-throughput sequencing, deep learning models,
and experimental validation provides a viable idea to systematically study the role of RNA-binding proteins. For example, how do RBPs regulate gene expression during zebrafish development?
3. Technical applications
Based on previous work, we have identified a large number of conserved RNA structural elements related to RNA translation, RNA degradation, etc. These RNA structures can be exploited to design
RNA molecules that better help the vaccine and RNA targeting drug development. Moreover, the understanding of RNA structure and the application of artificial intelligence can help us to design better
gRNA in the RNA therapy field.